

超高速视频级原子力显微镜—HS-AFM
- HS-AFM
- RIBM
- 国外国外
- 三个月内
- 按需定制
- 议价
- 2022-11-08 16:43:33
北京佰司特科技有限责任公司
- 英文名称
- High Speed AFM
超高速视频级原子力显微镜—HS-AFM
原子力显微镜(Atomic Force Microscope,AFM),一种可用来研究包括绝缘体在内的固体材料表面结构的分析仪器。它通过检测待测样品表面和一个微型力敏感元件之间的极微弱的原子间相互作用力来研究物质的表面结构及性质。将一对微弱力极端敏感的微悬臂一端固定,另一端的微小针尖接近样品,这时它将与其相互作用,作用力将使得微悬臂发生形变或运动状态发生变化。扫描样品时,利用传感器检测这些变化,就可获得作用力分布信息,从而以纳米级分辨率获得表面形貌结构信息及表面粗糙度信息。原子力显微镜可以测量材料物理性质、力学性能、磁学性能、热学性能、电学性能等方面的一些特征信息,但在扫描成像速度上一直存在局限性,太慢的扫描速度导致原子力显微镜无法捕捉到分子间的相互作用过程和一些快速的分子动态变化。
超高速视频级原子力显微镜(High-Speed Atomic Force Microscope,HS-AFM)由日本 Kanazawa 大学 Prof. Ando 教授团队研发,日本RIBM公司(生体分子计测研究所株式会社,Research Institute of Biomolecule Metrology Co., Ltd)商业化的产品,可以达到视频级成像的商业化原子力显微镜。HS-AFM突破了传统原子力显微镜“扫描成像速慢”的限制,能够在液体环境下超快速动态成像,分辨率为纳米水平。样品无需特殊固定,不影响生物分子的活性,尤其适用于生物大分子互作动态观测。超高速视频级原子力显微镜HS-AFM主要有两种型号,SS-NEX样品扫描(Sample-Scanning HS-AFM)以及PS-NEX探针扫描(Probe-Scanning HS-AFM)。推出至今,全球已有100多位用户,发表 SCI 文章 300 余篇,包括Science, Nature, Cell 等顶级杂志。

相较于目前市场上的原子力显微镜成像设备,HS-AFM突破了 “扫描成像速慢”的限制,扫描速度高可达 20 frame/s,并且有 4 种扫描台可供选择。样品无需特殊固定染色,不影响生物分子的活性,尤其适用于生物大分子互作动态观测。液体环境下直接检测,超快速动态成像,分辨率为纳米水平。探针小,适用于生物样品;悬臂探针共振频率高,弹簧系数小,避免了对生物样品等的损伤。悬臂探针可自动漂移校准,适用于长时间观测。采用动态PID控制,高速扫描时仍可获得清晰的图像。XY轴分辨率2nm;Z轴分辨率0.5nm。
HS-AFM不仅拥有超高扫描速率与原子级别分辨率,而且具有操作的简易性,使得对单分子动态过程的捕捉变得十分方便,为科研工作者研所和理解生物物理、生物化学、分子生物学、病毒学以及生物医学等领域的单分子动态过程提供了一款强大的工具。
全新的HS-AFM采用了新的高频微悬臂架构,更低噪音、更高稳定性的2控制器,高速扫描器,缓冲防震设计,主动阻尼,动态PID,驱动算法优化,多种前沿技术,可以实现在超高速下获取高分辨的生物样品信息。新系统整合了基于工作流程的操作软件,直观的用户界面与流程化、自动化的设置使得研究人员可以专注于实验设计,不需要复杂的操作和条件设置,快速获取数据,加速研究的产出。

日本RIBM公司的超高速视频级原子力显微镜HS-AFM的创新点:
★ 高频微悬臂
弹性系数: 0.1 N/m
曲率半径: <10 nm
共振频率: 400-600kHz in liquid
★ 高速扫描台
20 frames/s. (Standard scanner)
★ 缓冲防震+主动阻尼+动态PID+算法优化
缓冲防震
主动阻尼
动态PID控制:可自动改变反馈增益,保证了HS-AFM在高速扫描条件下仍可获得清晰的图像
探针自动漂移校准,适用于长时间样品观测
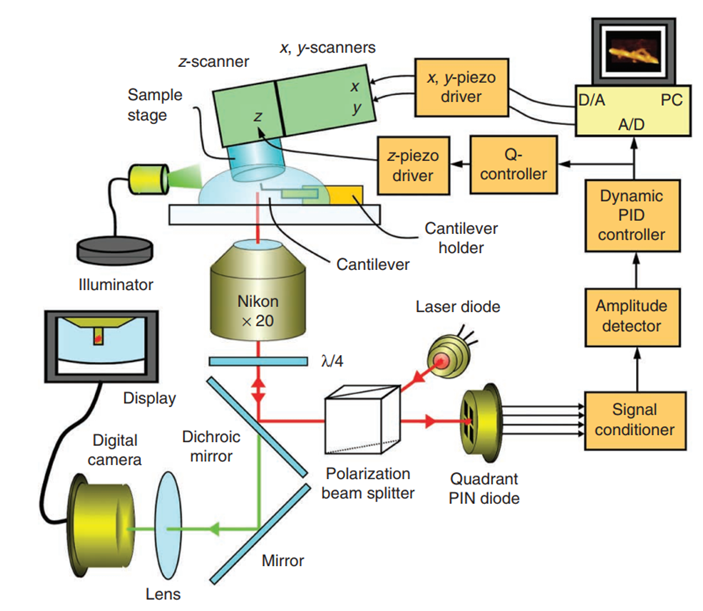
日本RIBM公司的超高速视频级原子力显微镜HS-AFM的的应用领域:
从单分子到单细胞,都可直接观测

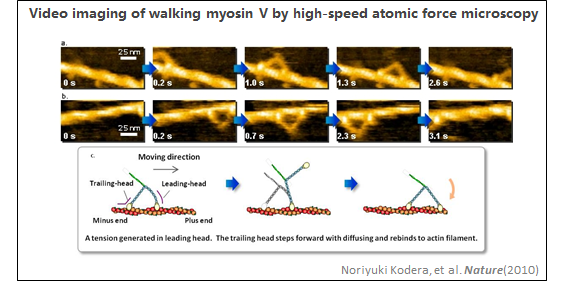
1、肌动蛋白
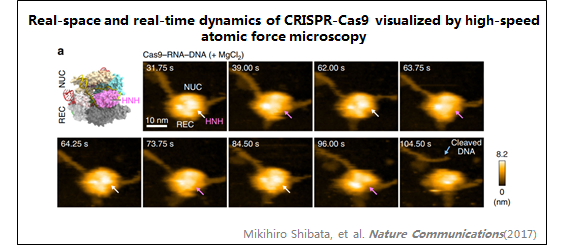
2、CRISPR-Cas9
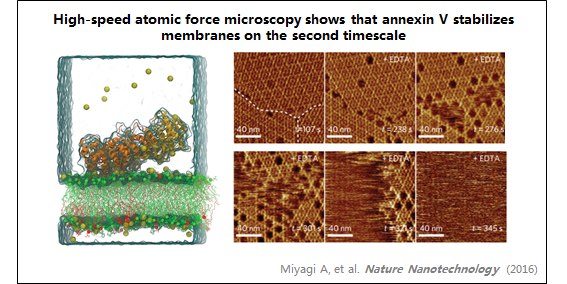
3、膜联蛋白
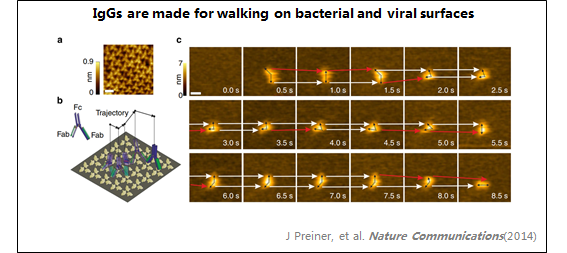
4、IgG
5、活细胞

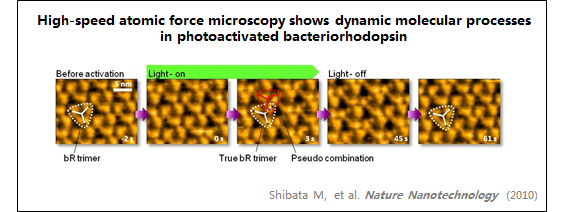
6、细菌视紫红质
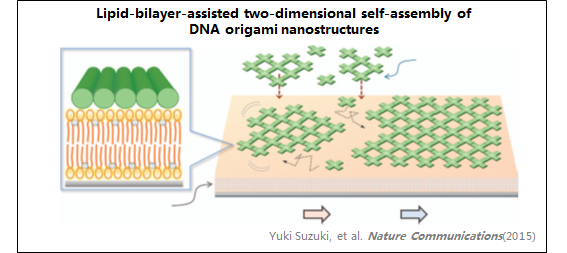
7、DNA纳米结构
8、仿生聚合物

日本RIBM公司的超高速视频级原子力显微镜HS-AFM的视频案例
1:IgG
在溶液中观察到抗体(IgG)。
IgG呈"Y"形,两个Fab区区分清晰。
由于锚定能力较弱,IgG保持其抗原结合能力。
2:Plasmid DNA
传统AFM在没有强锚定的情况下,DNA分子图像出现摆动。
然而,强锚定可能会削弱真实的结构和行为。
HS-AFM能清晰显示质粒的结构和运动,无强锚定。
3:DNA内切酶的消化:DNase I
DNA酶I是一种随机消化DNA的核酸内切酶。视频中的箭头表示DNase I消化DNA的部分。
请参考从DNA末端消化的核酸外切酶Bal31的视频。
4:DNA外切酶消化:Bal31
Bal31是一种从DNA链末端消化DNA的核酸外切酶。
视频显示Bal31的活性沿着DNA移动,并逐渐从DNA链的末端消化。
最后,DNA分子被完全消化,但环状DNA未被消化。高光点是Bal31分子,它们与DNA的不同位置结合。
5:DNA聚合酶的DNA延伸:Phi29
双链DNA(黄色)随着时间的推移而拉长。单链λDNA作为模具固定在基板上。
由于从随机六聚体引物(Red)结合到λDNA模体,phi29聚合酶(Black)以dNTP为底物合成互补DNA。
6:链亲和素2D晶体中的点缺陷
成功地观察到点缺陷在晶体中的扩散。
从图像上看,两个单空位缺陷的轨迹跟踪相对于晶格的两个轴是明显的各向异性的。
日本RIBM公司的超高速视频级原子力显微镜HS-AFM的文献列表
| Title | Journal | ||||
| Biophysical reviews top five: atomic force microscopy in biophysics | Biophysical Reviews | ||||
| Reconstruction of Three-Dimensional Conformations of Bacterial ClpB from High-Speed Atomic-Force-Microscopy Images |
Frontiers in Molecular Biosciences |
||||
| A facile combinatorial approach to construct a ratiometric fluorescent sensor: application for the real-time sensing of cellular pH changes |
Chemical Science | ||||
| DNA Nanotechnology to Disclose Molecular Events at the Nanoscale and Mesoscale Levels |
Springer Nature | ||||
| Quantitative description of a contractile macromolecular machine | Science Advances | ||||
| Dynamic Assembly/Disassembly of Staphylococcus aureus FtsZ Visualized by High-Speed Atomic Force Microscopy |
International Journal of Molecular Sciences 2021, Vol. 22, Page 1697 |
||||
| Localization atomic force microscopy | Nature 2021 594:7863 | ||||
| Movements of mycoplasma mobile gliding machinery detected by high-speed atomic force microscopy |
mBio | ||||
| An ultra-wide scanner for large-area high-speed atomic force microscopy with megapixel resolution |
Scientific Reports 2021 11:1 | ||||
| A molecularly engineered, broad-spectrum anti-coronavirus lectin inhibits SARS-CoV-2 and MERS-CoV infection in vivo |
Research Square | ||||
| Influenza virus ribonucleoprotein complex formation occurs in the nucleolus |
bioRxiv | ||||
| Tardigrade Secretory-Abundant Heat-Soluble Protein Has a Flexible β-Barrel Structure in Solution and Keeps This Structure in Dehydration |
Journal of Physical Chemistry B | ||||
| Ultrastructure of influenza virus ribonucleoprotein complexes during viral RNA synthesis |
Communications Biology on | ||||
| A facile combinatorial approach to construct a ratiometric fluorescent sensor: application for the real-time sensing of cellular pH changes |
Chemical Science | ||||
| Deformation of microtubules regulates translocation dynamics of kinesin |
Science Advances | ||||
| Unraveling the host-selective toxic interaction of cassiicolin with lipid membranes and its cytotoxicity |
bioRxiv | ||||
| Nanostructure and thermoresponsiveness of poly( N -isopropyl methacrylamide)-based hydrogel microspheres prepared via aqueous free radical precipitation polymerization |
RSC Advances | ||||
| JRAB/MICAL-L2 undergoes liquid–liquid phase separation to form tubular recycling endosomes |
Communications Biology 2021 4:1 |
||||
| Correlation of membrane protein conformational and functional dynamics |
Nature Communications 2021 12:1 |
||||
| Non-close-packed arrangement of soft elastomer microspheres on solid substrates |
RSC Advances | ||||
| Folding RNA–Protein Complex into Designed Nanostructures | Methods in Molecular Biology | ||||
| A glutamine sensor that directly activates TORC1 | Communications Biology 2021 4:1 |
||||
| Architecture of zero-latency ultrafast amplitude detector for high- speed atomic force microscopy |
Applied Physics Letters | ||||
| Correlative AFM and fluorescence imaging demonstrate nanoscale membrane remodeling and ring-like and tubular structure formation by septins |
Nanoscale | ||||
| Desiccation-induced fibrous condensation of CAHS protein from an anhydrobiotic tardigrade |
bioRxiv | ||||
| Construction of ferritin hydrogels utilizing subunit–subunit interactions |
PLOS ONE | ||||
| Monomeric α-synuclein (αS) inhibits amyloidogenesis of human prion protein (hPrP) by forming a stable αS-hPrP hetero-dimer. |
Prion | ||||
| Influence of protein adsorption on aggregation in prefilled syringes | Journal of Pharmaceutical Sciences |
||||
| Dynamic mechanisms of CRISPR interference by Escherichia coli CRISPR-Cas3 |
bioRxiv | ||||
| An RNA Triangle with Six Ribozyme Units Can Promote a Trans- Splicing Reaction through Trimerization of Unit Ribozyme Dimers |
Applied Sciences | ||||
| Faster high-speed atomic force microscopy for imaging of biomolecular processes Review of Scientific Instruments ARTICLE scitation.org/journal/rsi Faster high-speed atomic force microscopy for imaging of biomolecular processes |
Rev. Sci. Instrum | ||||
| Title: Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites |
bioRxiv | ||||
| Structural and dynamics analysis of intrinsically disordered proteins by high-speed atomic force microscopy |
Nature Nanotechnology | ||||
| Millisecond Conformational Dynamics of Skeletal Myosin II Power Stroke Studied by High-Speed Atomic Force Microscopy |
ACS Nano | ||||
| High-Speed Atomic Force Microscopy Reveals Spatiotemporal Dynamics of Histone Protein H2A Involution by DNA Inchworming |
The Journal of Physical Chemistry Letters |
||||
| Nanostructures, Thermoresponsiveness, and Assembly Mechanism of Hydrogel Microspheres during Aqueous Free-Radical Precipitation Polymerization |
Langmuir | ||||
| Chained structure of dimeric F 1-like ATPase in Mycoplasma mobile gliding machinery 4 |
bioRxiv | ||||
| Lipid Membrane Interaction of Peptide/DNA Complexes Designed for Gene Delivery |
Langmuir | ||||
| High-Speed Atomic Force Microscopy to Study Myosin Motility | Advances in Experimental Medicine and Biology |
||||
| Single-molecule level dynamic observation of disassembly of the apo-ferritin cage in solution |
Physical Chemistry Chemical Physics |
||||
| Atomic Force Microscopy of Biomembranes : A Tool for Studying the Dynamic Behavior of Membrane Proteins |
New Techniques for Studying Biomembranes |
||||
| Adenosine leakage from perforin-burst extracellular vesicles inhibits perforin secretion by cytotoxic T-lymphocytes |
PLOS ONE | ||||
| High-Speed Atomic Force Microscopy Reveals the Structural Dynamics of the Amyloid-β and Amylin Aggregation Pathways |
International Journal of Molecular Sciences 2020, Vol. 21, Page 4287 |
||||
| Molecular mechanism of the recognition of bacterially cleaved immunoglobulin by the immune regulatory receptor LILRA2 |
Journal of Biological Chemistry | ||||
| Biological physics by high-speed atomic force microscopy | Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences |
||||
| Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes |
Nucleic Acids Research | ||||
| Key Nucleation Stages and Associated Molecular Determinants and Processes in pH-Induced Formation of Amyloid Beta Oligomers as Revealed by High-Speed AFM |
bioRxiv | ||||
| Novel Babesia bovis exported proteins that modify properties of infected red blood cells |
PLoS Pathogens | ||||
| DNA origami demonstrate the unique stimulatory power of single pMHCs as T-cell antigens |
Proceedings of the National Academy of Sciences |
||||
| Structural insights into the mechanism of rhodopsin phosphodiesterase |
Nature Communications | ||||
| Structural insights into the mechanism of rhodopsin phosphodiesterase |
Nature Communications | ||||
| High-Speed AFM Reveals Molecular Dynamics of Human Influenza A Hemagglutinin and Its Interaction with Exosomes |
Nano Letters | ||||
| DNA Ring Motif with Flexible Joints | Micromachines | ||||
| Nanopores: a versatile tool to study protein dynamics | Essays in Biochemistry | ||||
| Geometrical Characterization of Glass Nanopipettes with Sub-10 nm Pore Diameter by Transmission Electron Microscopy |
Analytical Chemistry | ||||
| Carbon nanotube porin diffusion in mixed composition supported lipid bilayers |
Scientific Reports | ||||
| One-Step Calibration of AFM in Liquid | Frontiers in Physics | ||||
| Nanoscale interaction of RecG with mobile fork DNA | Nanoscale Advances | ||||
| Convergent evolution of processivity in bacterial and fungal cellulases |
Proceedings of the National Academy of Sciences |
||||
| Convergent evolution of processivity in bacterial and fungal cellulases |
Proceedings of the National Academy of Sciences of the United States of America |
||||
| Interaction of the motor protein SecA and the bacterial protein translocation channel SecYEG in the absence of ATP |
Nanoscale Advances | ||||
| Nanoreporter of an Enzymatic Suicide Inactivation Pathway | Nano Letters | ||||
| High-speed atomic force microscopy highlights new molecular mechanism of daptomycin action |
Nature Communications | ||||
| Direct visualization of the conformational change of FUS/TLS upon binding to promoter-associated non-coding RNA |
Chemical Communications | ||||
| Thermoresponsive Micellar Assembly Constructed from a Hexameric Hemoprotein Modified with Poly(N-isopropylacrylamide) toward an Artificial Light-Harvesting System |
Journal of the American Chemical Society |
||||
| Schizorhodopsins: A family of rhodopsins from Asgard archaea that function as light-driven inward H + pumps |
Science Advances | ||||
| Two-State Exchange Dynamics in Membrane-Embedded Oligosaccharyltransferase Observed in Real-Time by High-Speed AFM |
Journal of Molecular Biology | ||||
| Thermoresponsive structural changes of single poly(N-isopropyl acrylamide) hydrogel microspheres under densely packed conditions on a solid substrate |
Polymer Journal | ||||
| High-Speed Atomic Force Microscopy Reveals Factors Affecting the Processivity of Chitinases during Interfacial Enzymatic Hydrolysis of Crystalline Chitin |
ACS Catalysis | ||||
| Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge |
Nature Communications | ||||
| Assembly mechanism of a supramolecular MS-ring complex to initiate bacterial flagellar biogenesis in vibrio species |
Journal of Bacteriology | ||||
| Structural Dynamics of a Protein Domain Relevant to the Water- Oxidizing Complex in Photosystem II as Visualized by High-Speed Atomic Force Microscopy |
Journal of Physical Chemistry B | ||||
| Recent advances in bioimaging with high-speed atomic force microscopy |
Biophysical Reviews | ||||
| Dynamic behavior of an artificial protein needle contacting a membrane observed by high-speed atomic force microscopy |
Nanoscale | ||||
| Enhanced enzymatic activity exerted by a packed assembly of a single type of enzyme |
Chemical Science | ||||
| High Speed AFM and NanoInfrared Spectroscopy Investigation of A β1–42 Peptide Variants and Their Interaction With POPC/SM/Chol/GM1 Model Membranes |
Frontiers in Molecular Biosciences |
||||
| The hierarchical assembly of septins revealed by high-speed AFM | Nature Communications | ||||
| Millisecond dynamics of an unlabeled amino acid transporter | Nature Communications | ||||
| Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion |
Nature Structural & Molecular Biology |
||||
| A Simplified Cluster Analysis of Electron Track Structure for Estimating Complex DNA Damage Yields |
International Journal of Molecular Sciences |
||||
| Supramolecular tholos-like architecture constituted by archaeal proteins without functional annotation |
Scientific Reports | ||||
| Studies on the impellers generating force in muscle | Biophysical Reviews | ||||
| Diversity of physical properties of bacterial extracellular membrane vesicles revealed through atomic force microscopy phase imaging |
Nanoscale | ||||
| High-Speed AFM Reveals Molecular Dynamics of Human Influenza A Hemagglutinin and Its Interaction with Exosomes |
Nano Letters | ||||
| DNA density-dependent uptake of DNA origami-based two-or three- dimensional nanostructures by immune cells |
Nanoscale | ||||
| Spatiotemporally tracking of nano-biofilaments inside the nuclear pore complex core |
Biomaterials | ||||
| High-speed atomic force microscopy directly visualizes conformational dynamics of the HIV Vif protein in complex with three host proteins |
Journal of Biological Chemistry | ||||
| Self- and Cross-Seeding on α-Synuclein Fibril Growth Kinetics and Structure Observed by High-Speed Atomic Force Microscopy |
ACS Nano | ||||
| Liquidity Is a Critical Determinant for Selective Autophagy of Protein Condensates |
Molecular Cell | ||||
| Capturing transient antibody conformations with DNA origami epitopes |
Nature Communications | ||||
| Biophysics in Kanazawa University | Biophysical Reviews | ||||
| Dynamics of oligomer and amyloid fibril formation by yeast prion Sup35 observed by high-speed atomic force microscopy |
Proceedings of the National Academy of Sciences |
||||
| Annexin-V stabilizes membrane defects by inducing lipid phase transition |
Nature Communications | ||||
| Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity |
Science Advances | ||||
| Atomic Force Microscopy Visualizes Mobility of Photosynthetic Proteins in Grana Thylakoid Membranes |
Biophysical Journal | ||||
| Construction of a Hexameric Hemoprotein Sheet and Direct Observation of Dynamic Processes of Its Formation |
Chemistry Letters | ||||
| Single-molecule imaging analysis reveals the mechanism of a high- catalytic-activity mutant of chitinase A from Serratia marcescens |
Journal of Biological Chemistry | ||||
| Direct observation and analysis of TET-mediated oxidation processes in a DNA origami nanochip |
Nucleic Acids Research | ||||
| On-membrane dynamic interplay between anti-GM1 IgG antibodies and complement component C1q |
International Journal of Molecular Sciences |
||||
| Zwitterionic Polypeptides: Chemoenzymatic Synthesis and Loosening Function for Cellulose Crystals |
Biomacromolecules | ||||









